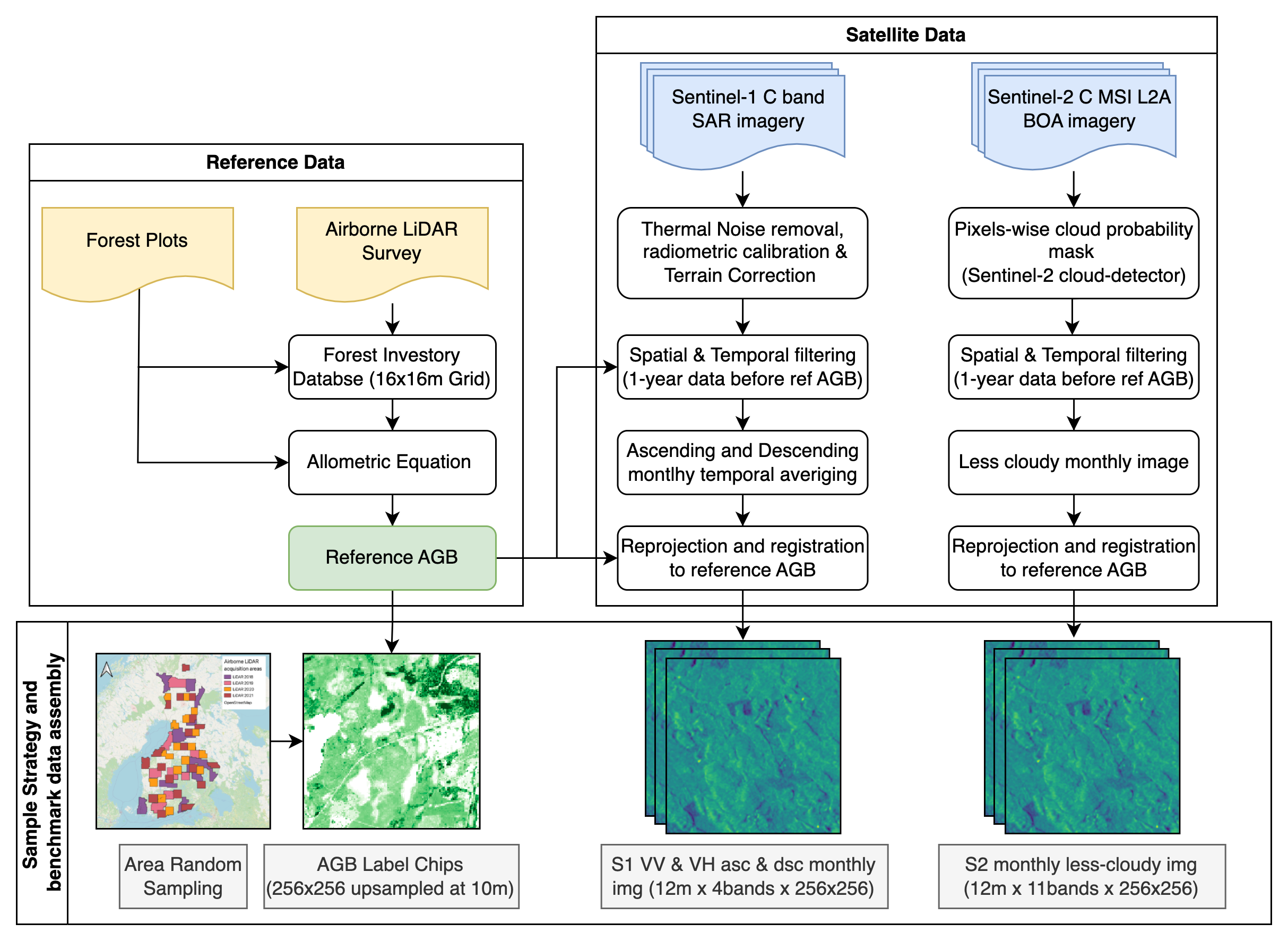
Data Collection Methods
The feature data for this dataset is imagery collected by the Sentinel-1 and Sentinel-2 satellite missions for nearly 13,000 patches of forest in Finland. Each patch (also called a "chip") represents a different 2,560 by 2,560 meter area of forest. The data were collected over a period of 5 years between 2016 and 2021. Each label in this challenge represents a specific chip, or a distinct area of forest. LiDAR measurements are used to generate the biomass label for each pixel in the chip. For each chip, a full year's worth of monthly satellite images for that area are provided, from the previous September to the most recent August. For example, for a LiDAR-based reference label chip from 2020, monthly satellite data is provided from September 2019 to August 2020. All of the satellite images have been geometrically and radiometrically corrected and resized to 10 meter resolution. Each resulting image is 256 by 256 pixels, and each pixel represents 10 square meters. Images represent monthly aggregations and are provided as GeoTIFFs with any associated geolocation data removed. You only need to generate one biomass prediction per chip, but can use as many of the chip's multi-temporal (different months) or multi-modal (Sentinel-1 or Sentinel-2) satellite images as you like. Predictions should include a yearly peak AGB value for each 10 by 10 pixel in the chip.
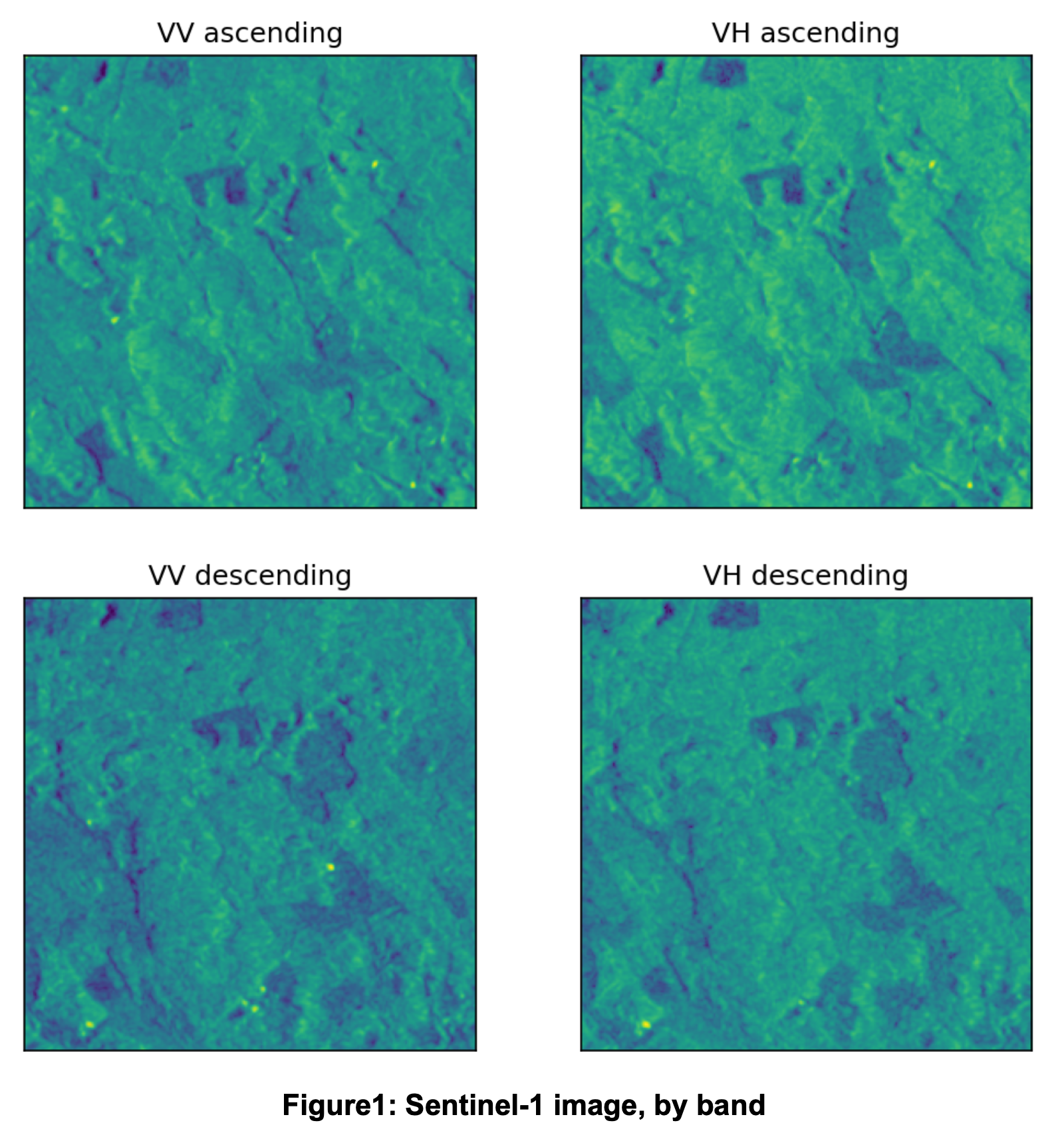
Example of a Sentinel-1 image: 001b0634_S1_00.tif is an image from Sentinel-1 provided as a part of the training dataset. The filename follows the format {chip_id}_{satellite}_{month_number}.tif, so we know that the chip_id is 001b0634 and that the image was captured by Sentinel-1 in September (the month number corresponds to the number of months since September).